SaTScan requires several different
types of files for analysis: 1) A case file with a column for the geographic unit. day, month or year (see documentation), and number of cases. You can aggregate the data into any geographic unit--large or small. 2) A geographic coordinate file (cartesian or lat/long) with the name of the unit (i.e. census tract), x and y for centroids of the geographic units, and 3) population file with the estimated population over the time period-- by year.
In this post, I will describe creating a case file using code in
R. The goal is to create a sum of homicides by month, year (just 2013 for this example), and police beat/post. We won't worry about any other specifics (i.e. degree) or related types of crimes, i.e. shootings.
To ready yourself for data preparation, read
Richard Block's tutorial or the more extensive
SatScan manual.
I use crime data from
Chicago's Open Data Portal. The same code can be applied to other types of data, health data, etc. A few key points: 1) the data contains victim-based data--which we want to convert into incidents. 2) not every post has a homicide, and 3) the reference post list contains 275 post. So, we will end up with a data set with 3300 rows (275 x 12 months) or simply a row for each post-month.
If you want to skip ahead and just look at the code, go to:
http://goo.gl/pmOi1u.
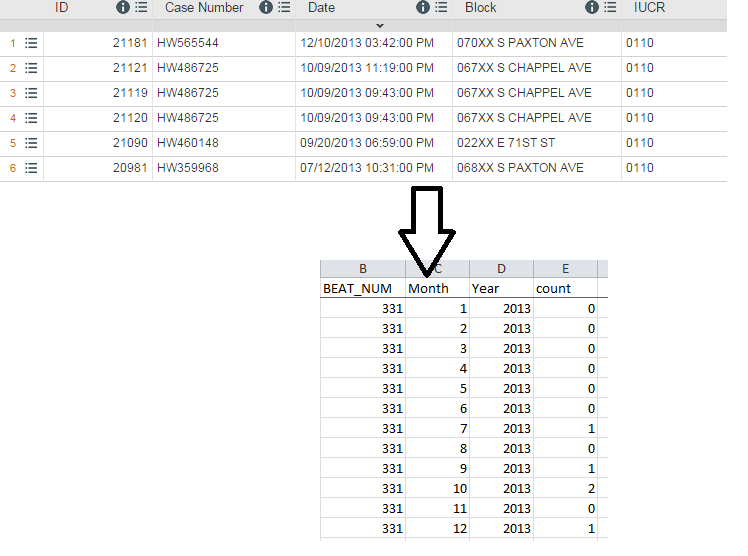 |
| At the top: What you start with. Bottom: After processing in R |
Overview of Steps: See the code for further details
Step #1: Two files are imported: 1) a victim-based file of all crimes, which is narrowed down to just homicides (you could also add in shootings) and 2) a 'reference' file or simply a list of the police beats/posts in Chicago.
Step #2: The data are summed up so that each row contains the total number of victims, then grouped again into incidents by using two different count variables.
Step #3: The list of police beats get column variables for each month in the year and expanded by reshaping data from wide to long. This serves as a 'reference list' for matching purposes.
Step #4: The two data sets are matched the 'unmatched' records are also kept. These are post-months that don't have a homicide, so each count value is replaced with a zero.
Step #5: To ensure the code has worked, I check the total number of rows (3300) and spot check various posts to make sure the data has been grouped in to incidents and posts correctly.
Whether in R or using for-fee software (i.e. SAS, STATA), preparing data for SaTScan is relatively straightforward but there are a number of steps.
Update #1 (2/18/15)
Scan statistics can also be implemented in R's Spatial Epi Package and rsatscan .



